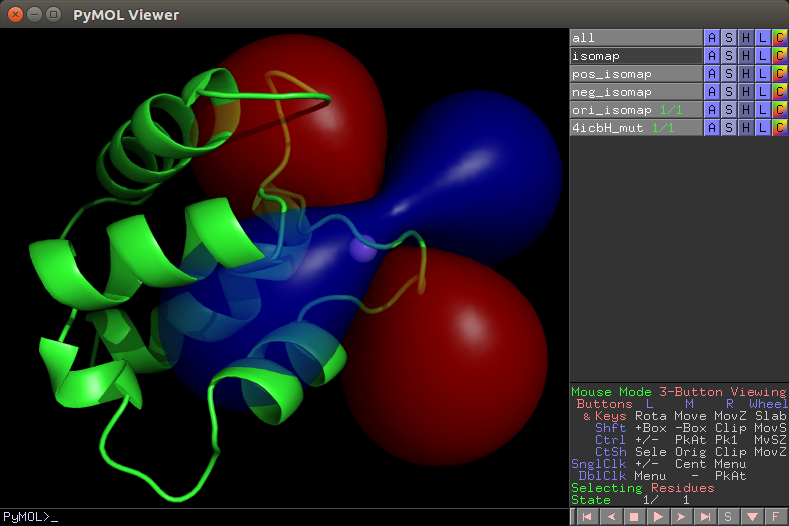
Plot PCS isosurface (PyMol view)¶
This example shows how to plot the PCS isosurface of a fitted \({\Delta\chi}\)-tensor for data from the example Fit Tensor to PCS Data. The isosurface can be viewed in PyMol.
Downloads¶
Download the data files
4icbH_mut.pdbandcalbindin_Er_HN_PCS_tensor.txtfrom here:Download the script pcs_plot_isosurface.py
Explanation¶
The protein and tensor are loaded as described previously in.
The isosurface files are generated using the function paramagpy.metal.Metal.isomap(). The contour level can be chosen by setting the isoval argument. A larger density value will result in a smoother surface. This function writes two files isomap.pml and isomap.pml.ccp4 which are the PyMol script and PCS grid files respectively.
The isosurface can be displayed by executing pymol isomap.pml from a terminal, or by selecting File>Run and navigating to the script isomap.pml.
Script¶
from paramagpy import protein, fit, dataparse, metal
# Load the PDB file
prot = protein.load_pdb('../data_files/4icbH_mut.pdb')
# Load the fitted tensor
met = metal.load_tensor('../data_files/calbindin_Er_HN_PCS_tensor.txt')
# Plot the isosurface to be opened in PyMol
met.isomap(prot.id, density=1, isoval=1.0)
Output¶
PyMol view of isosurface